HW2 必做题
Blast sequences
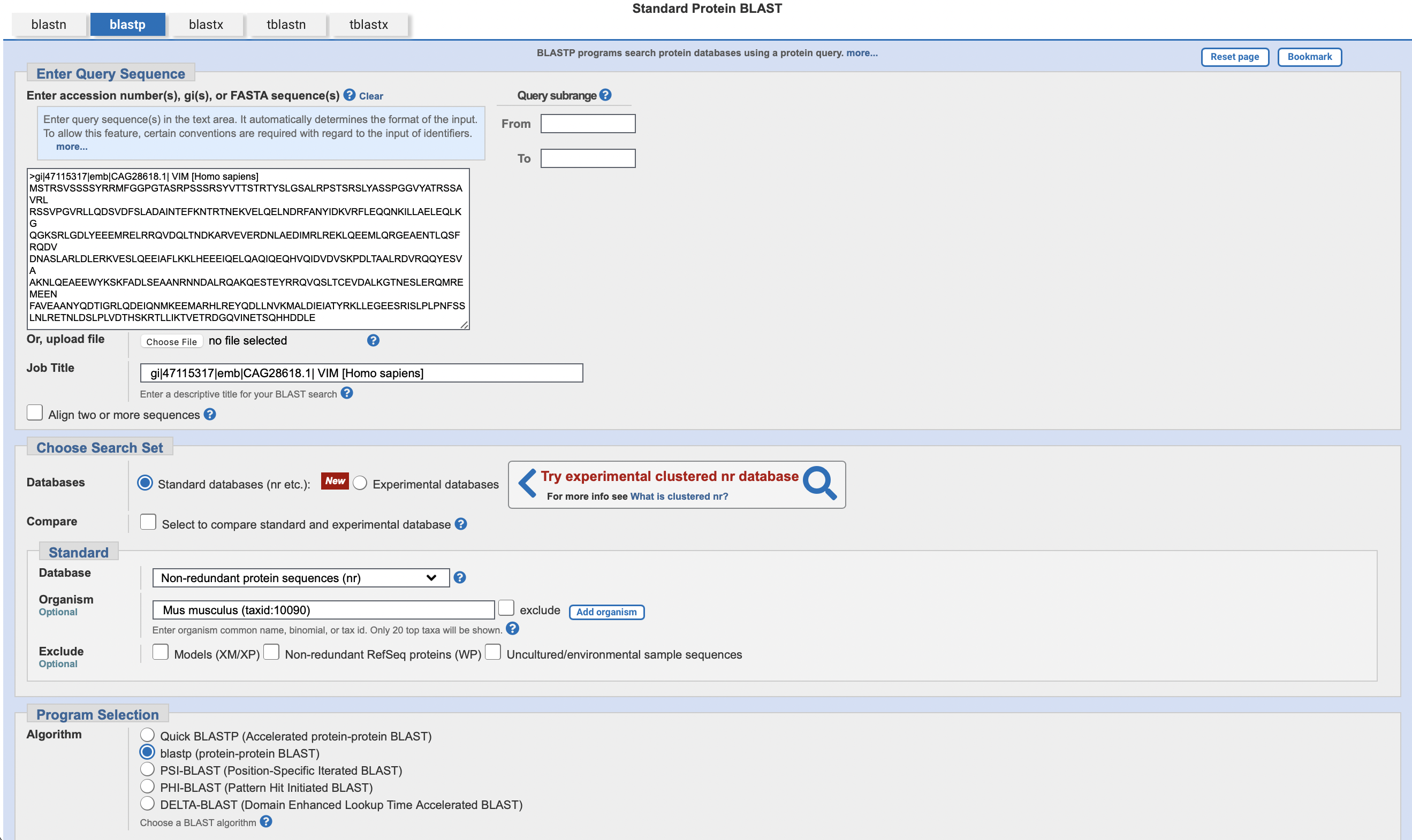
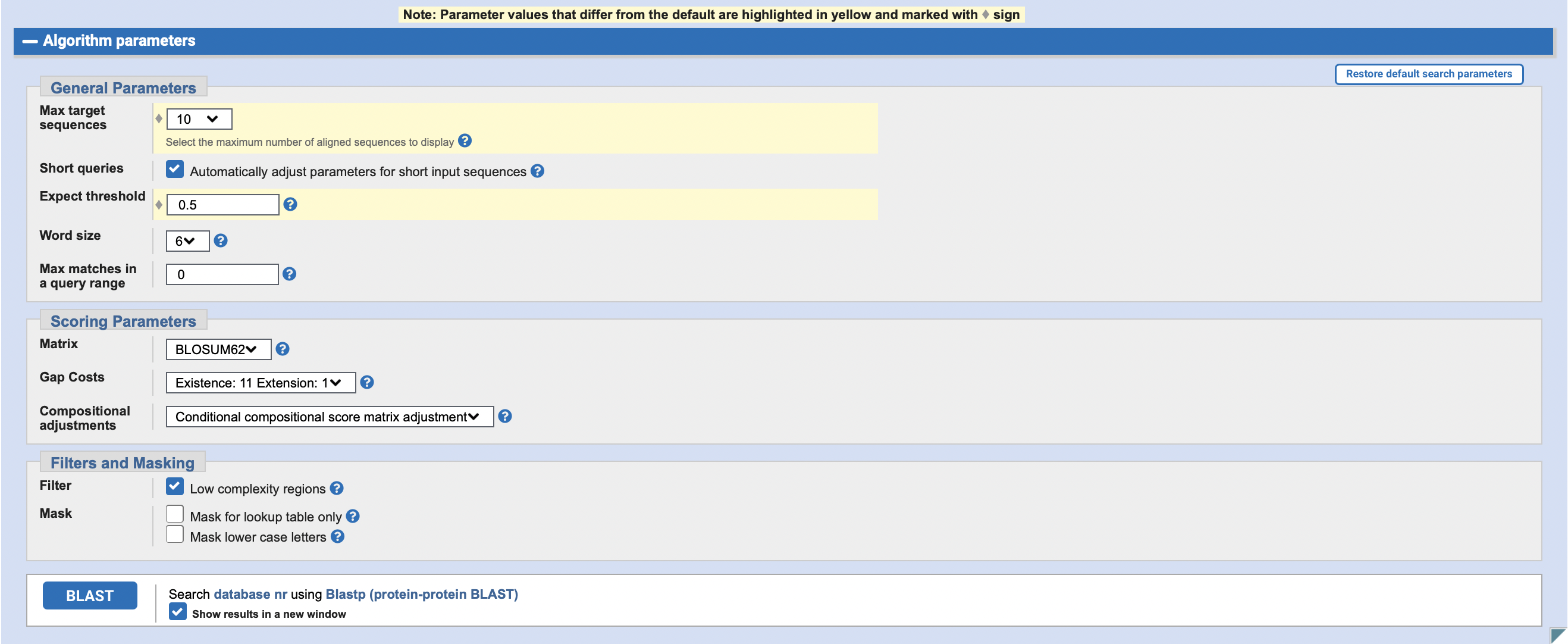
请使用网页版的 blastp, 将上面的蛋白序列只与 mouse protein database 进行比对, 设置输出结果最多保留10个, E 值最大为 0.5。将操作过程和结果截图。
注: blast的网站会提供多个mouse的databases,可以任选1个进行比对;也可以重复几次,每次选一个不同的database看看不同的输出结果,可以在作业中比较和讨论一下输出结果不同的原因。
操作:
![Screen Shot 2022-10-05 at 2.20.59 AM]()
![Screen Shot 2022-10-05 at 2.21.28 AM]()
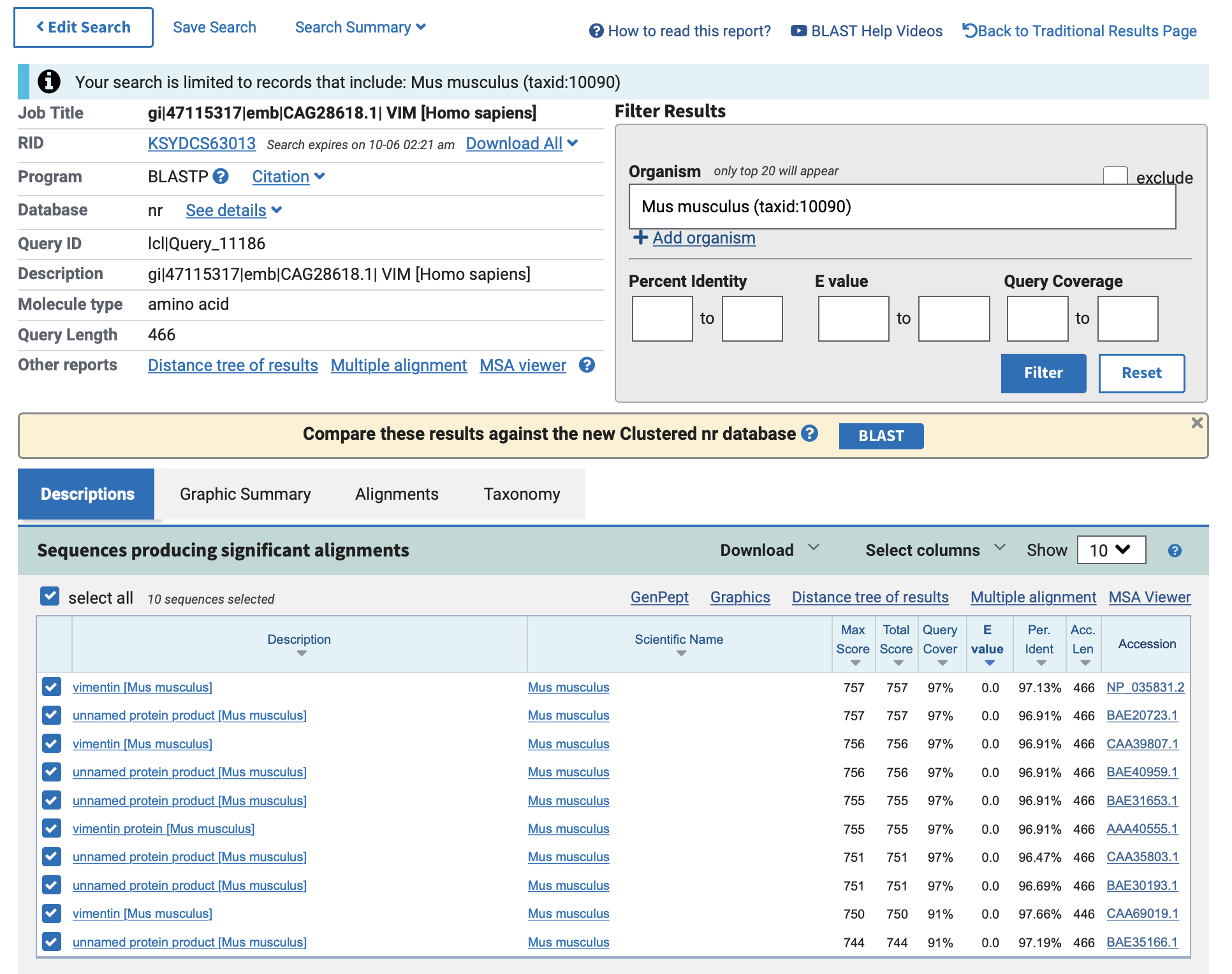
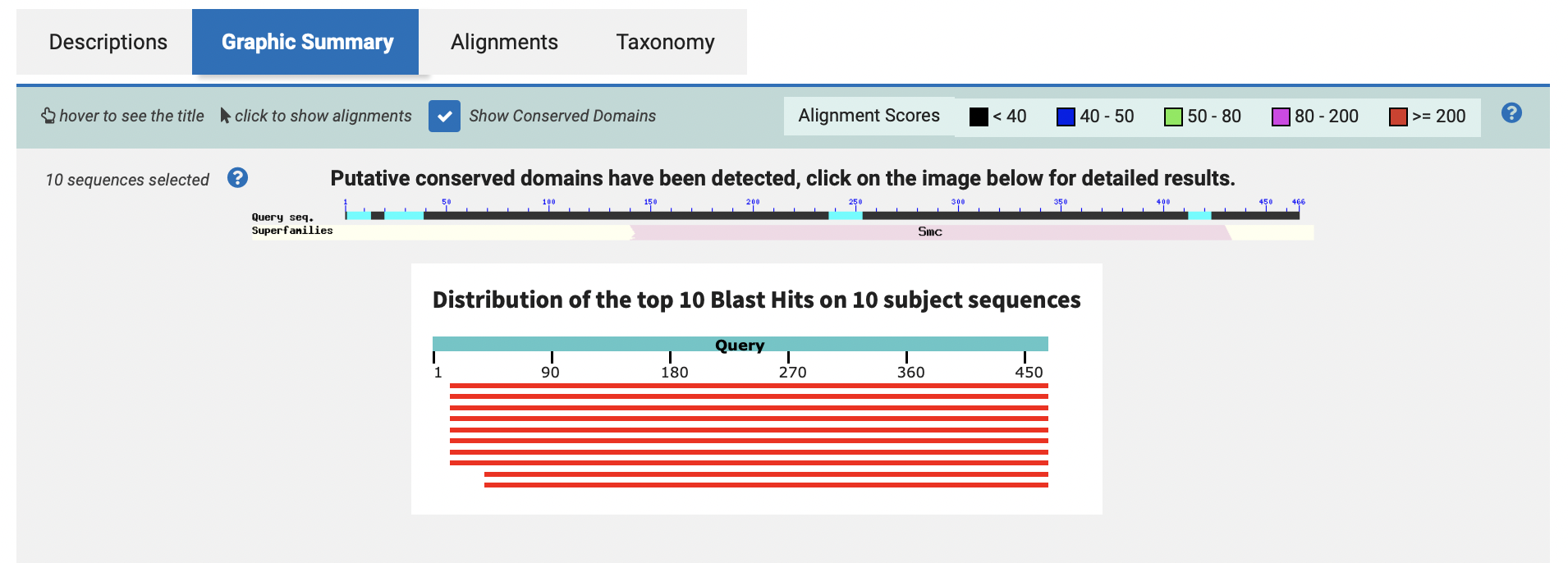
结果:
![Screen Shot 2022-10-05 at 2.23.09 AM]()
![Screen Shot 2022-10-05 at 2.23.26 AM]()
请使用 Bash 脚本编程:将上面的蛋白序列随机打乱生成10个, 然后对这10个序列两两之间进行 blast 比对,输出并解释结果。(请上传bash脚本,注意做好重要code的注释;同时上传一个结果文件用来示例程序输出的结果以及你对这些结果的解释。)
完整的代码文件以及生成的结果文件存放在仓库中,和该报告中的内容相同(仓库中多出clone文件夹中的10条随机序列文件、output文件夹中的所有blastp比对结果文件)。
bash脚本:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113
#!/bin/bash # 将原序列文件protein/VIM.fasta的内容读出到header, protein_string中 i=0 while read line; do if [ "$i" -eq 0 ]; then header="$line" else protein_string+="$line" fi i=$((i+1)) done < protein/VIM.fasta # 将protein_string字符串按字符存到protein_array数组中 while read -n 1 char ; do protein_array+=($char) done <<< "$protein_string" # 打乱函数(就地洗牌) function shuffle { local i tmp size rand size=${#protein_array[*]} for ((i=size-1; i>0; i--)); do rand=$(( $RANDOM % (i + 1) )) # swap tmp=${protein_array[i]} protein_array[i]=${protein_array[rand]} protein_array[rand]=$tmp done } # 隔70个字符就换行(fasta文件序列输出美观) function endline { local i size remainer size=${#protein_array[*]} for ((i=0; i<size; i++)); do new_protein_string+="${protein_array[$i]}" remainer=$(( $i % 70 )) if [ $remainer == 69 ]; then new_protein_string+="\n" fi done } [ ! -d clone ] && mkdir clone # 随机序列生成在clone文件夹中 total=10 # 生成的随机序列个数为10个 # 将原蛋白序列随机打乱生成total=10个,并存储到fasta文件中 for ((i=0; i<$total; i++)); do # 将原蛋白序列随机打乱 shuffle new_header=">CLONE$i" new_protein_string="" output="clone/clone$i.fasta" # 按fasta格式要求存储到文件 endline echo "${new_header}" > "$output" echo -e "${new_protein_string}" >> "$output" done # 用blastp进行序列比对,并将结果生成到文件 function blast { query="clone/clone$1.fasta" subject="clone/clone$2.fasta" output_blastp="output/blastp_$1_$2" blastp -query $query -subject $subject -out $output_blastp # 核心blastp命令 } # 读取并解析blastp生成的结果文件 function read_result { local i hit result_file valid_info score expect result_file="output/blastp_$1_$2" valid_info=`grep -A 3 "> CLONE$2" $result_file` # 获取"> CLONE$2"关键词往后3行内容 # 仅分析结果文件中的Score和Expect hit=0 # 记录query和subject序列是否有hit while read line; do if grep -q "Score" <<< "$line"; then hit=1 IFS=' ' read -r -a info_array <<< "$line" score=${info_array[2]} # Score IFS=',' read -r -a expect_array <<< "${info_array[7]}" expect=${expect_array[0]} # Expect fi i=$((i+1)) done <<< "$valid_info" # 输出比对的Score和Expect结果到result.txt文件中 if [ $hit -eq 1 ]; then echo "CLONE$1 vs CLONE$2: Score = $score, Expect = $expect" >> "result.txt" else echo "CLONE$1 vs CLONE$2: No hit" >> "result.txt" fi } # 对生成的total=10个随机序列两两之间进行blast比对并读取解析结果 for ((i=0; i<$total-1; i++)); do for ((j=i+1; j<$total; j++)); do blast $i $j # blast比对 read_result $i $j # 读取解析结果 done done
结果文件:
需要注意的是,运行bash脚本得到的文件分为三类:
- 10条随机序列:生成到
clone文件夹中,文件命名为clone/clone{id}.fasta,文件格式为fasta格式,header统一为>CLONE{id},id取值范围是0~9 - 10条序列两两比对的blastp结果:生成到
output文件夹中,文件命名为output/blastp_{query_id}_{subject_id},为blastp指令自动生成的结果文件,其中query_id和subject_id取值范围是0~9且二者不相等 - 汇总所有比对的简要结果:将blastp结果文件提取简要信息(最多一条alignment结果的Score和Expect)生成到
result.txt文件中
以下只显示第三种,即
result.txt文件:1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45
CLONE0 vs CLONE1: Score = 17.3, Expect = 1.1 CLONE0 vs CLONE2: Score = 15.0, Expect = 5.8 CLONE0 vs CLONE3: Score = 18.9, Expect = 0.33 CLONE0 vs CLONE4: Score = 17.3, Expect = 1.3 CLONE0 vs CLONE5: Score = 17.3, Expect = 1.1 CLONE0 vs CLONE6: Score = 16.9, Expect = 1.5 CLONE0 vs CLONE7: Score = 15.8, Expect = 3.8 CLONE0 vs CLONE8: Score = 21.9, Expect = 0.037 CLONE0 vs CLONE9: Score = 14.2, Expect = 9.3 CLONE1 vs CLONE2: Score = 19.6, Expect = 0.24 CLONE1 vs CLONE3: Score = 16.5, Expect = 1.9 CLONE1 vs CLONE4: Score = 16.9, Expect = 1.4 CLONE1 vs CLONE5: Score = 18.5, Expect = 0.44 CLONE1 vs CLONE6: Score = 16.5, Expect = 2.1 CLONE1 vs CLONE7: Score = 16.9, Expect = 1.3 CLONE1 vs CLONE8: Score = 16.2, Expect = 2.5 CLONE1 vs CLONE9: Score = 18.9, Expect = 0.38 CLONE2 vs CLONE3: Score = 15.4, Expect = 4.6 CLONE2 vs CLONE4: Score = 18.1, Expect = 0.69 CLONE2 vs CLONE5: Score = 17.3, Expect = 1.1 CLONE2 vs CLONE6: Score = 16.2, Expect = 2.7 CLONE2 vs CLONE7: Score = 19.2, Expect = 0.32 CLONE2 vs CLONE8: Score = 17.3, Expect = 1.1 CLONE2 vs CLONE9: Score = 17.3, Expect = 1.2 CLONE3 vs CLONE4: Score = 16.5, Expect = 2.3 CLONE3 vs CLONE5: Score = 15.0, Expect = 5.5 CLONE3 vs CLONE6: Score = 16.2, Expect = 2.5 CLONE3 vs CLONE7: Score = 18.5, Expect = 0.56 CLONE3 vs CLONE8: Score = 17.3, Expect = 1.0 CLONE3 vs CLONE9: Score = 16.2, Expect = 2.3 CLONE4 vs CLONE5: Score = 18.5, Expect = 0.49 CLONE4 vs CLONE6: No hit CLONE4 vs CLONE7: Score = 21.2, Expect = 0.071 CLONE4 vs CLONE8: Score = 18.1, Expect = 0.61 CLONE4 vs CLONE9: Score = 17.3, Expect = 1.1 CLONE5 vs CLONE6: Score = 16.2, Expect = 2.4 CLONE5 vs CLONE7: Score = 23.1, Expect = 0.019 CLONE5 vs CLONE8: Score = 15.0, Expect = 5.7 CLONE5 vs CLONE9: Score = 20.8, Expect = 0.10 CLONE6 vs CLONE7: Score = 17.3, Expect = 1.0 CLONE6 vs CLONE8: Score = 15.0, Expect = 6.1 CLONE6 vs CLONE9: Score = 17.3, Expect = 1.00 CLONE7 vs CLONE8: Score = 14.6, Expect = 8.6 CLONE7 vs CLONE9: Score = 18.1, Expect = 0.72 CLONE8 vs CLONE9: Score = 20.4, Expect = 0.12
对这些结果的解释:
其中将原蛋白序列随机打乱生成的10个新蛋白序列命名为CLONE0~CLONE9,且每两条序列间最多只取一组alignment的评分结果(取Score最高的一条)。对其比对的结果执行排序(优先按Score从大到小排,若Score相同则按Expect从小到大排):
1
sort -k6nr,6 -k9n,9 'result.txt'
得到排序后的结果:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45
CLONE5 vs CLONE7: Score = 23.1, Expect = 0.019 CLONE0 vs CLONE8: Score = 21.9, Expect = 0.037 CLONE4 vs CLONE7: Score = 21.2, Expect = 0.071 CLONE5 vs CLONE9: Score = 20.8, Expect = 0.10 CLONE8 vs CLONE9: Score = 20.4, Expect = 0.12 CLONE1 vs CLONE2: Score = 19.6, Expect = 0.24 CLONE2 vs CLONE7: Score = 19.2, Expect = 0.32 CLONE0 vs CLONE3: Score = 18.9, Expect = 0.33 CLONE1 vs CLONE9: Score = 18.9, Expect = 0.38 CLONE1 vs CLONE5: Score = 18.5, Expect = 0.44 CLONE4 vs CLONE5: Score = 18.5, Expect = 0.49 CLONE3 vs CLONE7: Score = 18.5, Expect = 0.56 CLONE4 vs CLONE8: Score = 18.1, Expect = 0.61 CLONE2 vs CLONE4: Score = 18.1, Expect = 0.69 CLONE7 vs CLONE9: Score = 18.1, Expect = 0.72 CLONE3 vs CLONE8: Score = 17.3, Expect = 1.0 CLONE6 vs CLONE7: Score = 17.3, Expect = 1.0 CLONE6 vs CLONE9: Score = 17.3, Expect = 1.00 CLONE0 vs CLONE1: Score = 17.3, Expect = 1.1 CLONE0 vs CLONE5: Score = 17.3, Expect = 1.1 CLONE2 vs CLONE5: Score = 17.3, Expect = 1.1 CLONE2 vs CLONE8: Score = 17.3, Expect = 1.1 CLONE4 vs CLONE9: Score = 17.3, Expect = 1.1 CLONE2 vs CLONE9: Score = 17.3, Expect = 1.2 CLONE0 vs CLONE4: Score = 17.3, Expect = 1.3 CLONE1 vs CLONE7: Score = 16.9, Expect = 1.3 CLONE1 vs CLONE4: Score = 16.9, Expect = 1.4 CLONE0 vs CLONE6: Score = 16.9, Expect = 1.5 CLONE1 vs CLONE3: Score = 16.5, Expect = 1.9 CLONE1 vs CLONE6: Score = 16.5, Expect = 2.1 CLONE3 vs CLONE4: Score = 16.5, Expect = 2.3 CLONE3 vs CLONE9: Score = 16.2, Expect = 2.3 CLONE5 vs CLONE6: Score = 16.2, Expect = 2.4 CLONE1 vs CLONE8: Score = 16.2, Expect = 2.5 CLONE3 vs CLONE6: Score = 16.2, Expect = 2.5 CLONE2 vs CLONE6: Score = 16.2, Expect = 2.7 CLONE0 vs CLONE7: Score = 15.8, Expect = 3.8 CLONE2 vs CLONE3: Score = 15.4, Expect = 4.6 CLONE3 vs CLONE5: Score = 15.0, Expect = 5.5 CLONE5 vs CLONE8: Score = 15.0, Expect = 5.7 CLONE0 vs CLONE2: Score = 15.0, Expect = 5.8 CLONE6 vs CLONE8: Score = 15.0, Expect = 6.1 CLONE7 vs CLONE8: Score = 14.6, Expect = 8.6 CLONE0 vs CLONE9: Score = 14.2, Expect = 9.3 CLONE4 vs CLONE6: No hit
可以看到CLONE5和CLONE7的Score = 23.1、Expect = 0.019比对结果最好。
更进一步查看二者比对的blastp结果文件output/blastp_5_7:
1 2 3 4 5 6
Score = 23.1 bits (48), Expect = 0.019, Method: Compositional matrix adjust. Identities = 12/32 (38%), Positives = 18/32 (56%), Gaps = 0/32 (0%) Query 256 SKQLEAPSNSSHIEVVSGRVSNLDYKGTNKRN 287 ++ LE S S I +SG + +L Y G +RN Sbjct 158 AQDLEVASQRSDINDLSGILEHLFYNGQMRRN 189可以看到CLONE5的第256~287处序列SKQLEAPSNSSHIEVVSGRVSNLDYKGTNKRN,和CLONE7的第158~189处序列AQDLEVASQRSDINDLSGILEHLFYNGQMRRN的详细比对结果(打分、E值、一致性、相似率、缺失/插入率,以及具体哪部分序列match或mismatch)。
- 10条随机序列:生成到